N-terminal Amino Acid Sequence Analysis by MALDI-TOF MS
N-terminal Amino Acid Sequence Analysis
N-terminal Amino Acid Sequence Analysis by MALDI-TOF MS
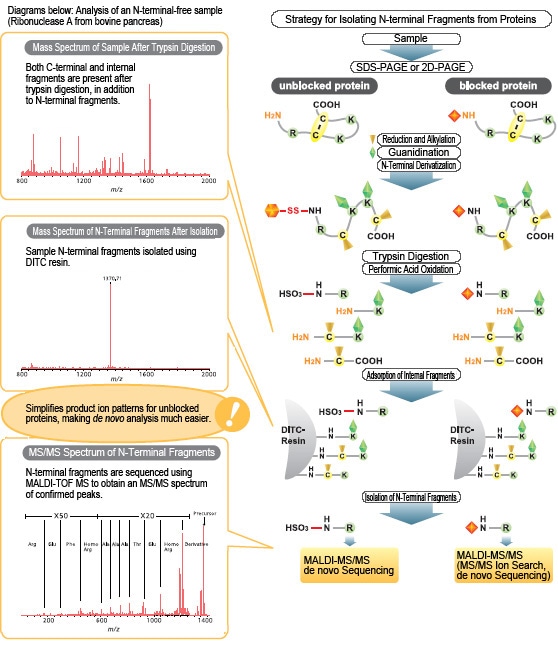
N-terminal fragments can be isolated even from proteins with blocked N-terminals. N-terminal fragments from proteins with unblocked N-terminals can be recovered as sulfonic acid derivatives, which permitsde novosequencing by MALDI-MS/MS. N-terminal fragments can be separately isolated even if multiple proteins coexist in a single SDS-PAGE sample (band).
ORFinder-NB Mass Sequencing Kit (Protein N-Terminal Sequencing Kit)
- Isolates N-terminal fragments from trypsin digests of proteins using DITC resins, whether the protein N-terminals are blocked or not.
- Recovers N-terminal fragments from proteins with unblocked N-terminals as sulfonic acid derivatives. As a result, primarily y-sequence ions are detected in MS/MS spectra, enablingde novosequence analysis.
ORFinder-NB Mass Sequencing Kit (Protein N-Terminal Sequencing Kit)

Permits isolation of N-terminal fragments, even from proteins with blocked N-terminals. N-terminal fragments from proteins with unblocked N-terminals can be recovered as sulfonic acid derivatives, which permitsde novosequencing by MALDI-MS/MS. N-terminal fragments can be separately isolated even if multiple proteins coexist in a single SDS-PAGE sample (band).
MALDI-TOF MS Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometer

MALDI-TOF MS AXIMA Performance is a TOF-TOF type MALDI-TOF MS featuring high-energy CID.
Achieves even higher MS/MS measurement sensitivity, offering high reliability for proteome analysis. Automated MS (or MS/MS) on multiple samples combined with Mascot database searches permits the rapid identification of candidate protein markers.


